workshops
Last Updates: 2024-10-17
Missing Values
is.na()
Missing values in R are coded as NA, a special value in R. When importing or tyding your data, it’s important to ensure that you’re properly identifying missing values, and handling them with the arguments na.strings, if using read.csv(), or na, if using read_csv().
na_values <- c("NA", "NULL", "", " ")
data_gapminder <- read.csv("../data/gapminder_nas.csv", na.strings = na_values)
With the data loaded, we can investigate missing values. is.na() returns a logical vector, TRUE for NA, otherwise FALSE. This is easiest with a single vector…
# get colnames
colnames(data_gapminder)
## [1] "country" "continent" "year" "lifeExp" "pop" "gdpPercap"
# generate logical vector
missing_pop <- is.na(data_gapminder$pop)
typeof(missing_pop)
## [1] "logical"
head(missing_pop)
## [1] FALSE FALSE FALSE FALSE FALSE FALSE
The logical vector on it’s own, is not super helpful. We can generate counts of the TRUE values by summing the number of TRUE instances in missing_pop…
sum(missing_pop)
## [1] 223
We can do this more interactively with
sum(is.na(data_gapminder$pop))
## [1] 223
Complete Cases
Another point of interest in looking at missing values is to explore the number of observations for which no data are missing, which can be done with complete.cases(). Similar to is.na(), complete.cases() returnds a logical vector…
complete_gapminder <- complete.cases(data_gapminder)
typeof(complete_gapminder)
## [1] "logical"
We can get a count, like before
sum(complete_gapminder)
## [1] 853
And for use later, when subsetting, we can also generate a vector of those which are complete, identified by their index number. This is done with which()
# first few index numbers of comlpete cases
head(which(complete_gapminder))
## [1] 2 3 4 5 8 12
Unique values & duplicates
unique(), returns a vector of unique values…
unique(data_gapminder$continent)
## [1] "Asia" "Europe" "Africa" "Americas" "Oceania"
duplicated() returns a logical vector, TRUE for values (of a vector) or rows (of a data frame) that are duplicates, FALSE if they are unique. Since the first instance of a value is unique, the first value will return FALSE, and any subsequent duplicates will return TRUE.
# on a vector
head(duplicated(data_gapminder$continent))
## [1] FALSE TRUE TRUE TRUE TRUE TRUE
# there are 5 unique continents and 1699 duplicates
sum(duplicated(data_gapminder$continent))
## [1] 1699
# on a data frame
head(duplicated(data_gapminder))
## [1] FALSE FALSE FALSE FALSE FALSE FALSE
# there are no duplicate values
sum(duplicated(data_gapminder))
## [1] 0
Descriptive Statistics
Basic stats fucntions in R generally do not remove NA values by default, and if your data has NA values, they will return
NA. You can avoid these errors withna.rm = TRUEas an argument.
Number of Observations
The number of values in a single vector can be derived with length()
length(data_gapminder$country)
## [1] 1704
When used on a data frame, however, length() returns the number of columns…
length(data_gapminder)
## [1] 6
When working with data frames, it is preferable to use nrow() and ncol() for the number of rows and columns, respectively.
ncol(data_gapminder)
## [1] 6
nrow(data_gapminder)
## [1] 1704
Range
The range of your data can be calculated with range().
range(data_gapminder$year, na.rm = TRUE)
## [1] 1952 2007
If you’re only interested in the min or max values individually, this can be pulled with min() and max() respectively.
min(data_gapminder$year, na.rm = TRUE)
## [1] 1952
max(data_gapminder$year, na.rm = TRUE)
## [1] 2007
Mean & Median
The mean and median are calculated with mean() and median() respectively…
mean(data_gapminder$lifeExp, na.rm = TRUE)
## [1] 58.9021
median(data_gapminder$gdpPercap, na.rm = TRUE)
## [1] 3242.531
Variance and Standard Deviation
The variance and SD are calculated with var() and sd()…
var(data_gapminder$lifeExp, na.rm = TRUE)
## [1] 165.1613
sd(data_gapminder$lifeExp, na.rm = TRUE)
## [1] 12.85151
Quantiles
Quantiles can be derived with quantile(), which defaults to the max, min and interquartiles.
quantile(data_gapminder$gdpPercap, na.rm = TRUE)
## 0% 25% 50% 75% 100%
## 241.1659 1147.3888 3242.5311 8533.0888 113523.1329
Summaries
Much of the above can be gathered all at once if needed with summary(). summary() can be run on a single vector, or on an entire data frame…
summary(data_gapminder$gdpPercap)
## Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
## 241.2 1147.4 3242.5 6839.5 8533.1 113523.1 219
summary(data_gapminder)
## country continent year lifeExp
## Length:1704 Length:1704 Min. :1952 Min. :23.60
## Class :character Class :character 1st Qu.:1962 1st Qu.:47.83
## Mode :character Mode :character Median :1977 Median :59.63
## Mean :1979 Mean :58.90
## 3rd Qu.:1992 3rd Qu.:70.60
## Max. :2007 Max. :82.60
## NA's :217 NA's :192
## pop gdpPercap
## Min. :6.001e+04 Min. : 241.2
## 1st Qu.:2.736e+06 1st Qu.: 1147.4
## Median :6.703e+06 Median : 3242.5
## Mean :2.835e+07 Mean : 6839.5
## 3rd Qu.:1.836e+07 3rd Qu.: 8533.1
## Max. :1.319e+09 Max. :113523.1
## NA's :223 NA's :219
A better summary would come from properly denoted data types…
# summary with the first three columns classed as categorical data
summary(data_gapminder)
## country continent year lifeExp
## Afghanistan: 12 Africa :624 1962 :132 Min. :23.60
## Albania : 12 Americas:300 1957 :128 1st Qu.:47.83
## Algeria : 12 Asia :396 1967 :127 Median :59.63
## Angola : 12 Europe :360 1952 :125 Mean :58.90
## Argentina : 12 Oceania : 24 1982 :125 3rd Qu.:70.60
## Australia : 12 (Other):850 Max. :82.60
## (Other) :1632 NA's :217 NA's :192
## pop gdpPercap
## Min. :6.001e+04 Min. : 241.2
## 1st Qu.:2.736e+06 1st Qu.: 1147.4
## Median :6.703e+06 Median : 3242.5
## Mean :2.835e+07 Mean : 6839.5
## 3rd Qu.:1.836e+07 3rd Qu.: 8533.1
## Max. :1.319e+09 Max. :113523.1
## NA's :223 NA's :219
Visualizing Distributions
Plotting for presentation will be covered in greater detail in a future session.
Basic plotting for distribution analysis is relatively straight forward in base R.
Distribution of a Single Variable
Histograms
You can plot a basic histogram with hist()
hist(data_gapminder$gdpPercap)
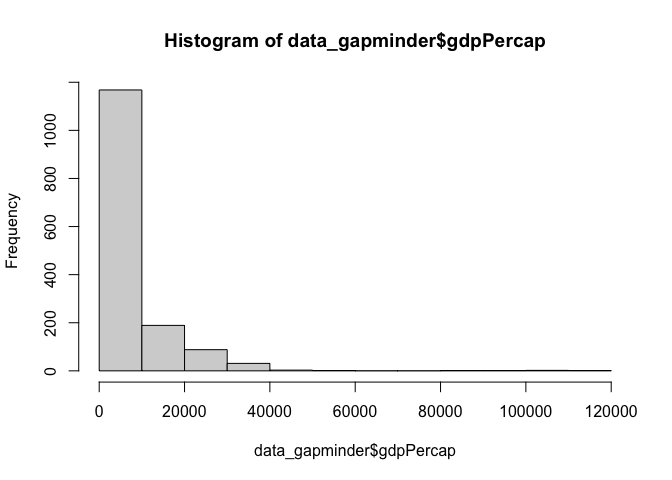
And you can increase the ‘buckets’ with the breaks argument
hist(data_gapminder$gdpPercap, breaks = 30)
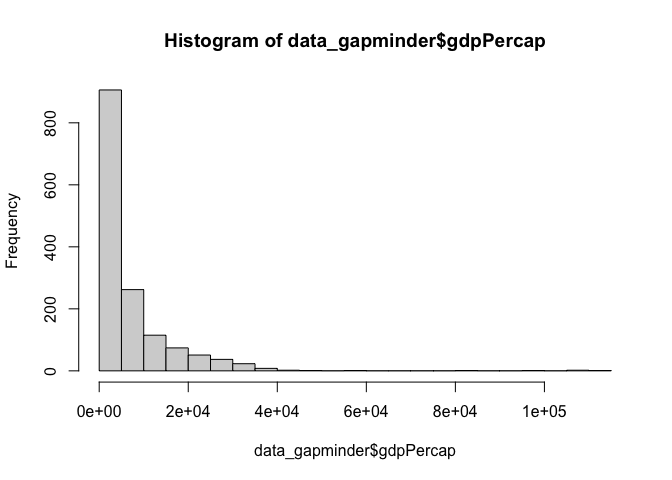
Density Plots
A density plot can be generated with plot(), but requires the extra step of computing the density of the data with the density() function first
dens <- density(data_gapminder$lifeExp, na.rm = TRUE)
plot(dens)
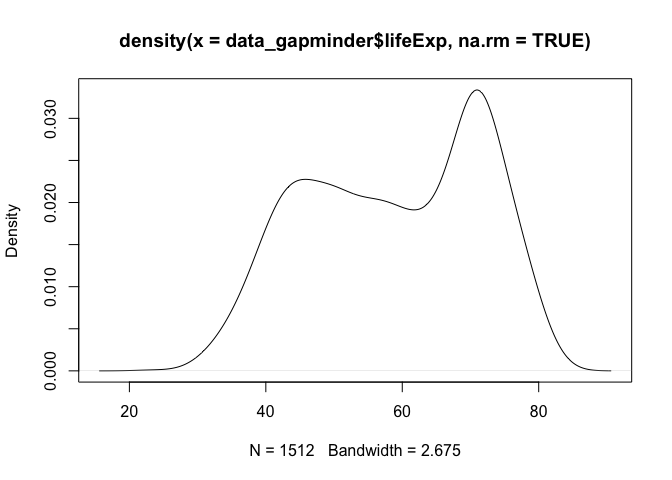
Distribution of Two Variables
Box Plots
boxplot() allows you to facet one variable by another using a tilde ~ to define the relationship
boxplot(data_gapminder$lifeExp ~ data_gapminder$continent)
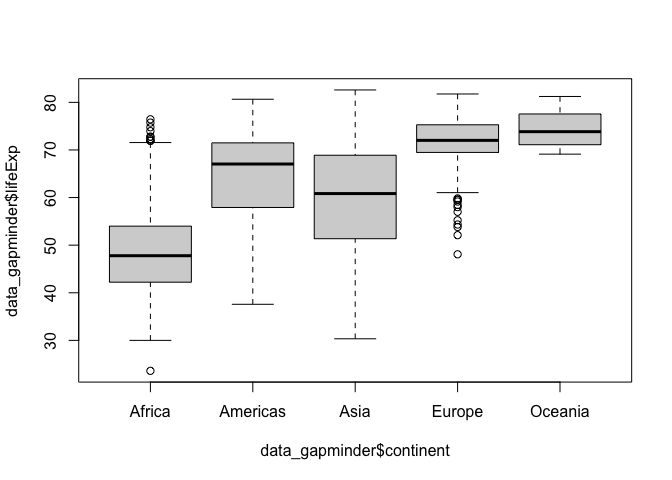
Scatter Plots
plot() by default assumes a single variable as in the earlier example, but will generate a scatter plot if provided with two variables, the first assigned to the x-axis, the second to the y-axis.
plot(x = data_gapminder$gdpPercap, y = data_gapminder$lifeExp)
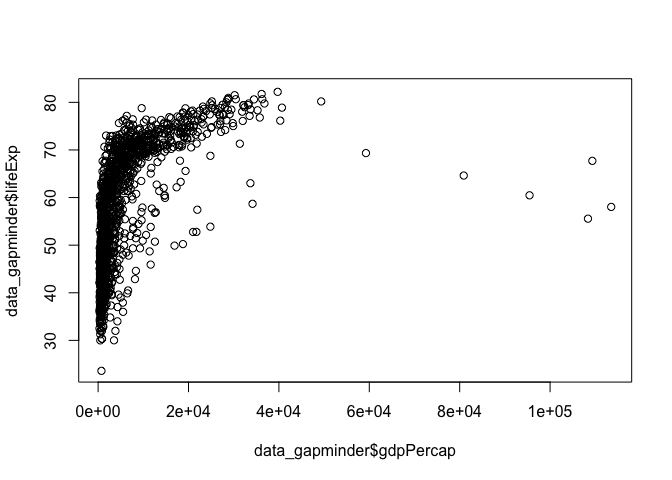
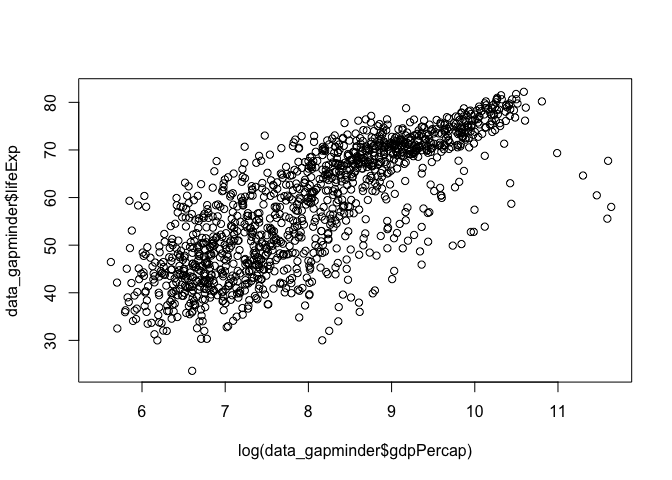
You can also transform you data within the function for quick investigation
# log transform gdp per capita
plot(x = log(data_gapminder$gdpPercap), y = data_gapminder$lifeExp)
Test your R might
Here are some examples for you to look at and challenge yourself, ensuring you now have a better understanding of today’s session!
Q1. You are given the following dataset of students’ grades:
df <- data.frame(
Name = c("John", "Alice", "Bob", "Alice", "Mary", "John"),
Math = c(85, NA, 79, NA, 93, 85),
Science = c(78, 88, 84, 88, NA, 78),
English = c(92, 90, 85, 90, 88, 92)
)
Part A. Identify the missing values and compute how many complete cases there are in this dataset.
Part B. How many duplicate rows exist, and how would you remove them?
Part C. Once you’ve removed the duplicates, compute the mean Math score, handling the missing values properly.
Q2. You have a dataset of car attributes, with columns: Horsepower, Weight, and MPG (Miles per Gallon) [To access the dataset, you can use data(mtcars)].
Part A. Calculate the summary statistics (mean, median, and standard deviation) for each variable.
Part B. Create a box plot to compare the distribution of Horsepower and MPG. What insights can you derive from the box plot?
Part C. Create a scatter plot between Weight and MPG. Explain whether there is any relationship between these two variables and interpret the plot.