In this part, we begin performing meta-analysis using the R language. If you are not familiar with R, you may refer to R for Data Science for a comprehensive introduction.
We will cover how to prepare data for import into RStudio, import the data, label it appropriately, and analyze it using meta-analysis codes.
Data Preparation
To perform meta-analysis, we first need to calculate the mean change and the Standard Deviation (SD) of the mean change for studies reporting only pre- and post-measurements. Additionally, we should label our data based on defined criteria (discussed further below) to prepare for subgroup analysis, which is critical in meta-analyses.
Let’s say we have extracted the following data from a study evaluating C-reactive protein (CRP) levels in blood after consuming Vitamin E (Ahmadi et al. 2013):
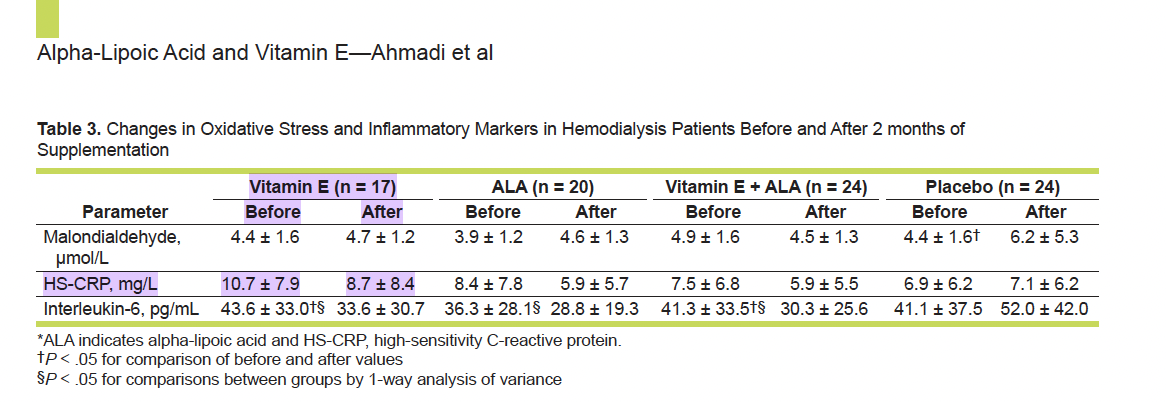
Our extracted data should look like this:
| Mean_Pre_CRP | SD_Pre_CRP | Mean_Post_CRP | SD_Post_CRP | Mean_Change_CRP | SD_Change_CRP |
|---|---|---|---|---|---|
| 10.7 | 7.9 | 8.7 | 8.4 | ? | ? |
Now we can calculate the mean change and SD of change using the following formulas:
- Mean Change = \( Mean_{Post} - Mean_{Pre} \)
- SD of Change = \( \sqrt{(SD_{Pre}^2 + SD_{Post}^2) - (2 \times r \times SD_{Pre} \times SD_{Post})} \)
The formula for calculating SD of Change requires a pre-post correlation (r). In general in RCTs, the Pre and Post Mean and SD, or Mean/SD changes are reported. However, if the latter are not, we need some way of estimating r. One way of doing this is to draw from an included study in which the authors do report r or SD of Change. If the SD of Change is reported, we can re-order the above formula to solve for r:
- r: \( \frac{SD_{Pre}^2 + SD_{Post}^2 - SD_{change}^2}{2 \times SD_{Pre} \times SD_{Post}} \)
In our example the SD of change is not reported. Therefore, to calculate r, we use the following known study information (Pirhadi-Tavandashti et al. 2020):
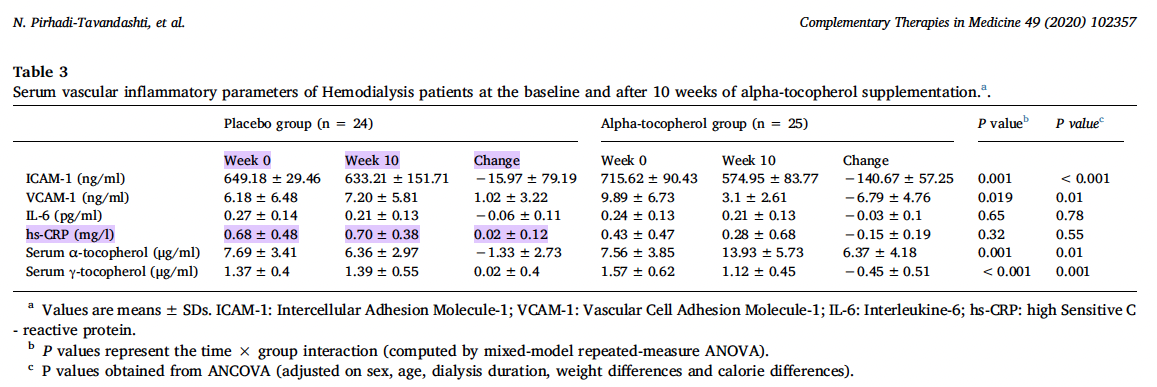
r: \( \frac{(0.48^2) + (0.38^2) - (0.12^2)}{2 \times 0.48 \times 0.38} = 0.98 \)
So in our example, we can assume r as 0.98.
SD change = \( \sqrt{((7.9^2) + (8.4^2)) - (2 \times 0.98 \times 7.9 \times 8.4)} = 1.70 \)
Considering these, we can now complete our table:
| Mean_Pre_CRP | SD_Pre_CRP | Mean_Post_CRP | SD_Post_CRP | Mean_Change_CRP | SD_Change_CRP |
|---|---|---|---|---|---|
| 10.7 | 7.9 | 8.7 | 8.4 | -2 | 1.70 |
Note: You can study more about pre-post correlation here.
Subgroup Analysis Preparation
To perform subgroup analysis, we must assign codes to variables based on which we want to conduct the analysis. Commonly used variables include:
- Gender
- Dosage
- Duration of intervention
- Health status of individuals
- Adjustment for baseline values
- Study design
- Adherence to the intervention
These variables can be coded as numeric values using mutate() function from dplyr package. For example, gender can be coded as: Both genders = Code 1, Male = Code 2, Female = Code 3.
For this part, we will use a sample data for demonstration. In this sample dataset, we want to understand how using Vitamin E affects the levels of CRP in blood. You can download the data to your local computer from Sample Data. The gender column indicates the gender of individuals researchers recruited in their study.
Let’s see how we can add a new column to our data, coding gender:
|
|
|
|
|
|
|
|
These codes facilitate subgroup analysis to understand how variables like gender affect outcomes. We will discuss the “how” in the next part.
Meta-Analysis
The first analysis we will become familiar with is the main meta-analysis, which we can perform in R using metacont from the meta package. We highly recommend running help(meta) as it provides comprehensive information on how to run your analyses and which codes to use.
Let’s see how we can do meta-analysis in our sample dataset. Again, in this sample dataset, we want to understand how using Vitamin E affects the levels of CRP in blood:
|
|
|
|
In this sample data, the effect of Vitamin E on blood CRP levels in adults were analyzed (this is demonstration-only data). The common effect model represents the result of pooling all studies together. The pooled estimate is -0.40 [-0.52; -0.27], which means that consuming Vitamin E may reduce CRP levels in the blood by approximately 0.40 mg/L. The next question is: Is this finding statistically significant? The numbers within brackets indicate the 95% confidence interval (CI). If this interval includes zero, the finding is not statistically significant. If it does not include zero, the finding is significant at the 95% confidence level (which we specified for this analysis). In our example, the confidence interval does not include zero, indicating that the result is statistically significant. The p-value further confirms this, being less than 0.0001.
Note: Usually, the first thing you need to check after performing a meta-analysis is the heterogeneity, as it indicates whether the model you have chosen for the analysis is appropriate.
Heterongenity is a very important concept in meta-anlysis studies. Heterogeneity refers to differences among studies. When heterogeneity is high or statistically significant, it means that the included studies differ considerably in certain characteristics (like methodology).
Note: The less variation there is between the studies (i.e., the lower the heterogeneity), the more valid and precise the findings from the meta-analysis will be.
Note: I^2 is a quantitative estimate of the proportion of variability among effect sizes due to heterogeneity. So technically, it is a statistical proxy for heterogeneity. It does not tell you why heterogeneity exists, just that it probably does. So that is why we will also try to look for the source of heterogeneity by comparing study designs, populations, dosages, follow-up durations, risk of bias, etc.
So how do we assess heterogeneity? We usually use the following criteria as a general rule of thumb:
-
Based on I-squared (I^2):
- Less than 50: Low heterogeneity
- Greater than 50: High heterogeneity
- Value of zero: No heterogeneity
-
Based on P-value for heterogeneity (from Cochran’s Q test):
- P-value less than 0.05 or 0.1 indicates significant heterogeneity
In our example, the I^2 is 66.8% and the p-value is 0.0003, indicating significant heterogeneity across studies. We can use I^2 as an indicator to help decide whether to use a fixed or random effects model.
-
Fixed Model: This model assumes that the studies are similar to each other or that there is low heterogeneity among them. In other words, selecting this model in the analysis causes the R to perform the meta-analysis under the assumption of low heterogeneity. Therefore, this model should be used when heterogeneity is low (I^2 below 50%).
-
Random Model:: This model assumes that the studies are different from each other or that there is high heterogeneity among them. In other words, selecting this model in the analysis causes the R to perform the meta-analysis under the assumption of high heterogeneity. Therefore, this model should be used when heterogeneity is high (I^2 above 50%).
In our example, since we have high heterogeneity, we should use a random effects model.
|
|
|
|
The results changed under the random model and are no longer statistically significant. In other words, Vitamin E consumption leads to a non-significant reduction in CRP levels by 0.43 mg/L. This analysis was conducted under the assumption that the studies are heterogeneous. Note that the level of heterogeneity remains similar in both the Fixed and Random models.
Visualization
The main way to visualize a meta-analysis is by using a forest plot. We will use the forest() function from the meta package, as shown in the code below:
|
|
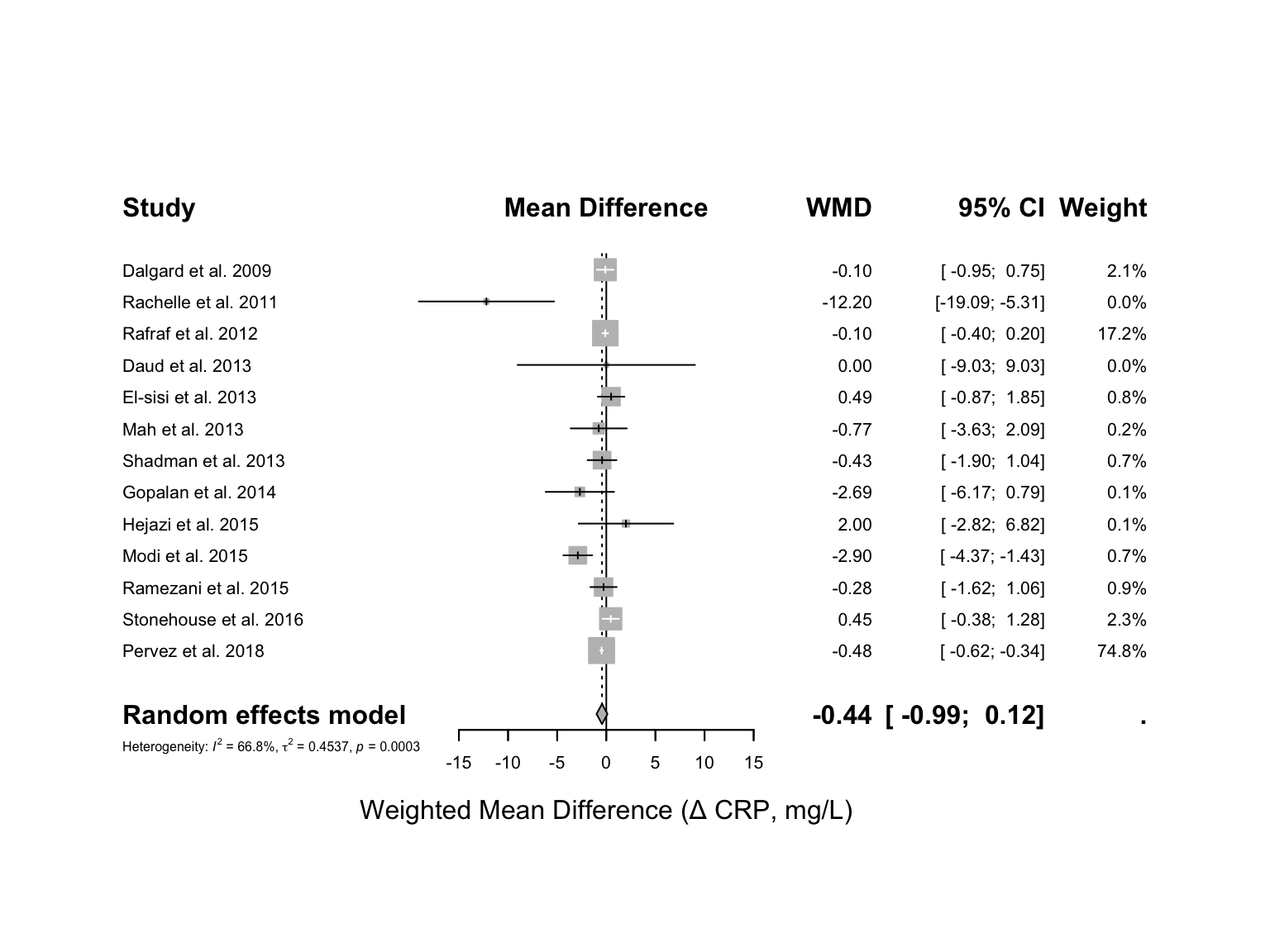
You can modify your plot using the various options offered by the forest() function. To see all available options, run ?forest in your console.
In the next part, we will explore finding the sources of heterogeneity, which will lead us to performing subgroup analyses.
Stay tuned!